Hey Ritu,
Welcome to the community, here are a couple of tips. For inactive batches (not generations) you are right that it defines the ones that will not be counted towards the final result, so if you want 50 counted batches you need to assign batches to 50+inactive batches. You are correct for the overall generations being a sum of gen/batch and # of batches, same as the # of total counted particles being #particles/gensgens/batch# of batches. I have found that generally you want gens/batches to be equal to # of batches for most sims, though there might be situations where it differs.
First thing you should do once you get your code running openmc.run is to run it in geometry debug mode like others suggested. First increase you particle amount to a higher amount (1 or 2 magnitudes higher usually) to make sure that every cell gets particles within it, and check that geometry debug doesn’t come up with any issues. Your inactive batches shouldn’t be arbitrary and you should run a simulation searching for Shannon entropy once you get your geometry and everything situated. Here are resources for that,
there is a section on Shannon entropy in this documentation, and here is an example of it,
It is easier than you might expect when first reading into it, you’re just plotting a graph and selecting where you think it becomes flat (converged) and that is your inactive batches. It will be given in generations so you will have to convert it back to batches.
For source definition you can just use a point source and use more inactive batches as the source will always adjust and converge to the actual source distribution as the transport runs continue,
# Create a point source
point = openmc.stats.Point((0, 0, 0))
source = openmc.IndependentSource(space=point)
settings = openmc.Settings()
settings.source = source
settings.batches = 100
settings.inactive = 10
settings.particles = 1000
settings.export_to_xml()
but if you want to do it correctly the following code will give you a simplistic source defined evenly over your geometry,
bounds = [-10, -10, -10, 10, 10, 10]
uniform_dist = openmc.stats.Box(bounds[:3], bounds[3:])
source = openmc.IndependentSource(space=uniform_dist)
settings.source = source
settings.batches = 100
settings.inactive = 10
settings.particles = 1000
settings.export_to_xml()
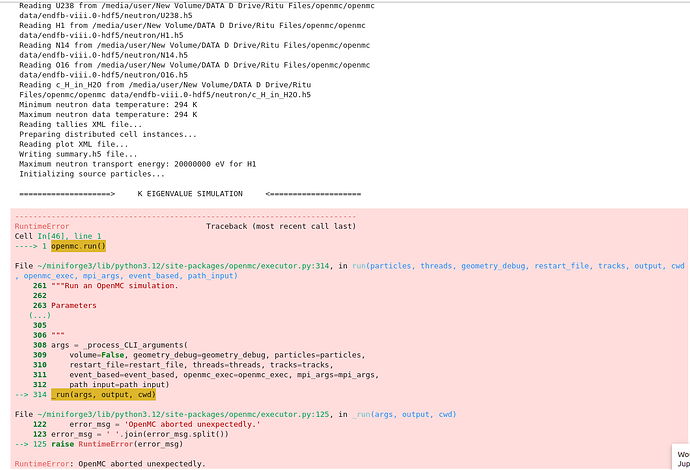
The bounds here are the lower and upper coordinates of a rectangular prism surrounding your geometry preferably only the section that includes fuel but that is not really necessary. Lastly for your error with OpenMC aborting unexpectedly, I can’t tell based on the code you have provided as of now, but I would expect it to be something fundamental like you’re not properly exporting an XML (Geometry, Settings, and Materials are required), or because your settings don’t actually work. I think this because it doesn’t give you a specific error and just aborts suddenly, which usually if its an error within either the material definition or geometry definition you get a specific error.